General organization
The genome viewer can be accessed through many pages on the Listeriomics website. From the home webpage, from the transcriptomics and proteomics summary pages, from the gene tool webpage, and from the small RNA tool webpage.
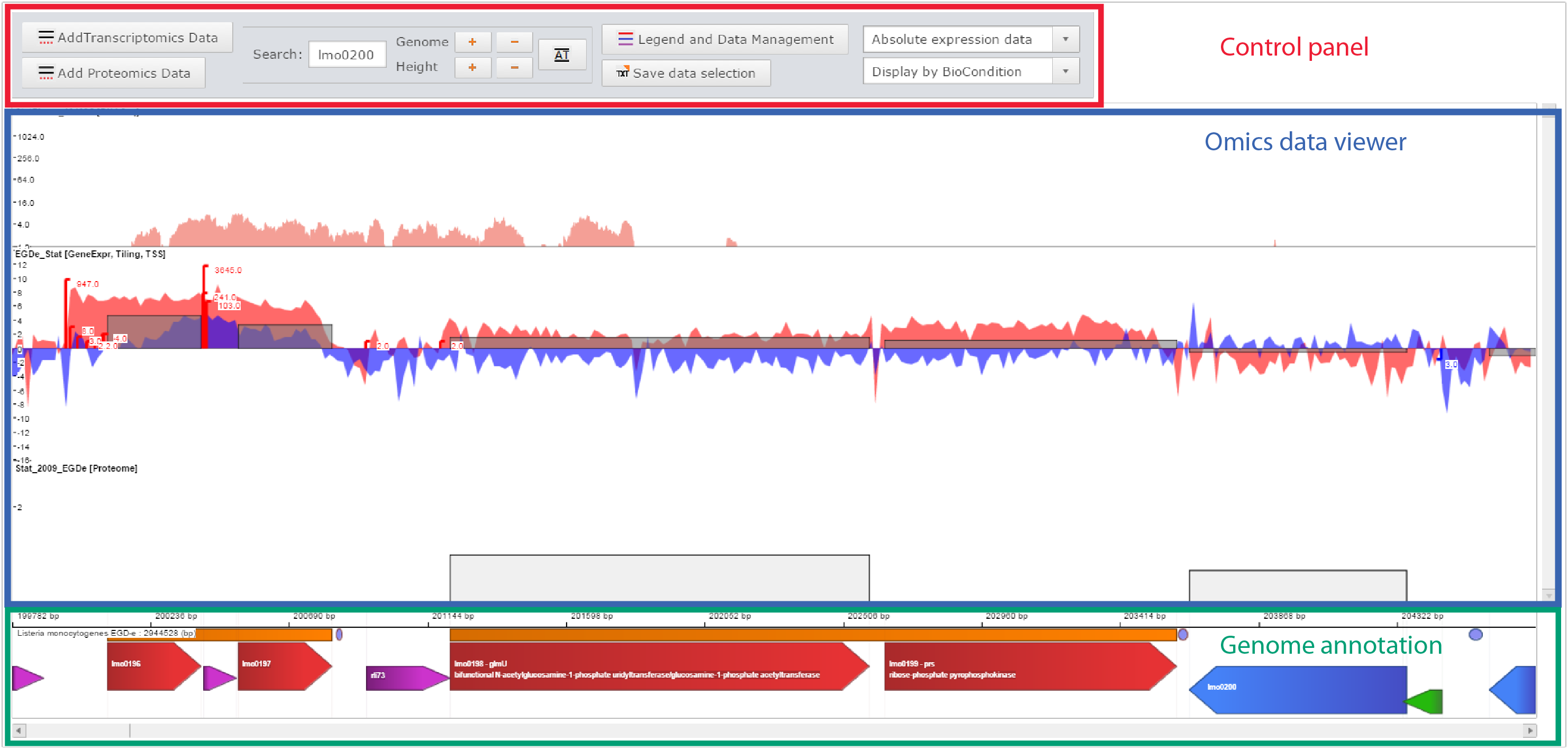 Printscreen of the Genome viewer of the Listeriomics website |
Control panel
- Add data
- Move the genome position, zoom in, zoom out
- Manage displayed datasets
Omics data viewer
The panel in which "omics" datasets are displayed
Genome annotation
All genome features available for the displayed Listeria strain genome.
|
Control panel
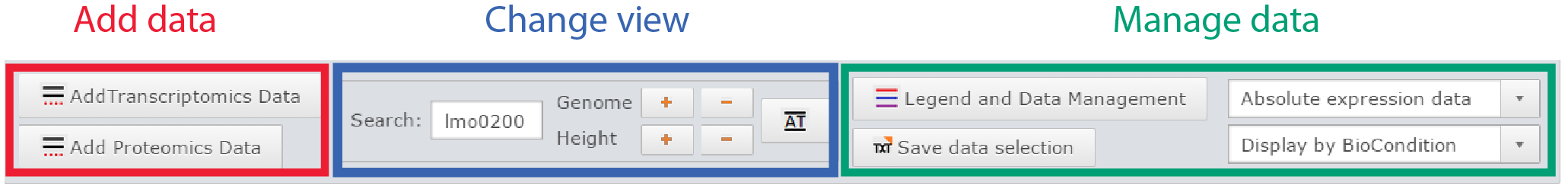
Control panel of the genome viewer
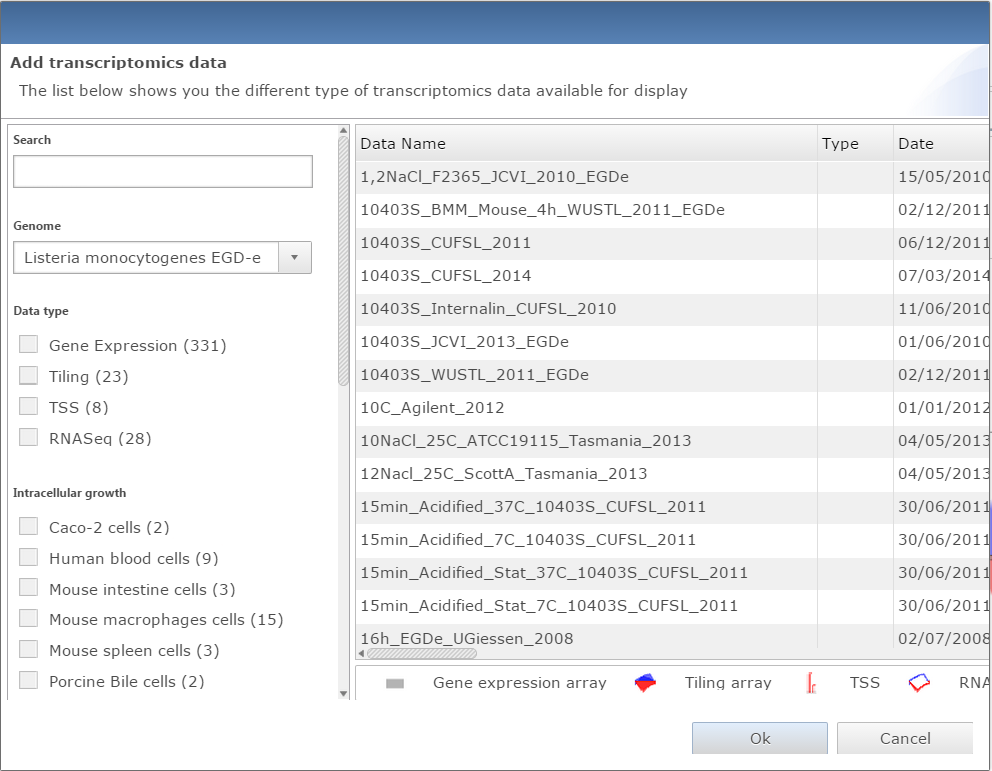 Dialog window for adding data to genome viewer |
Add data:
Two buttons are available to add datasets, one for transcriptomic data, the other for proteomic. Both will open a similar dialog with all datasets available. The user can filter datasets or directly search them. Every dataset selected will be displayed on the genome viewer by clicking on the Ok button.
|
 Printscreen of the same region with different bp scale |
Change view:
- Scroll the genome by searching a specific element contained in the displayed Listeria strain genome.
- Zoom in and zoom out horizontally, to change the base pair scale of display.
- AT button gives instant access to a view at the sequence scale where DNA sequence is displayed, along with amino acid sequence in the 6 possible frames.
- Increase the height of all genome tracks.
|
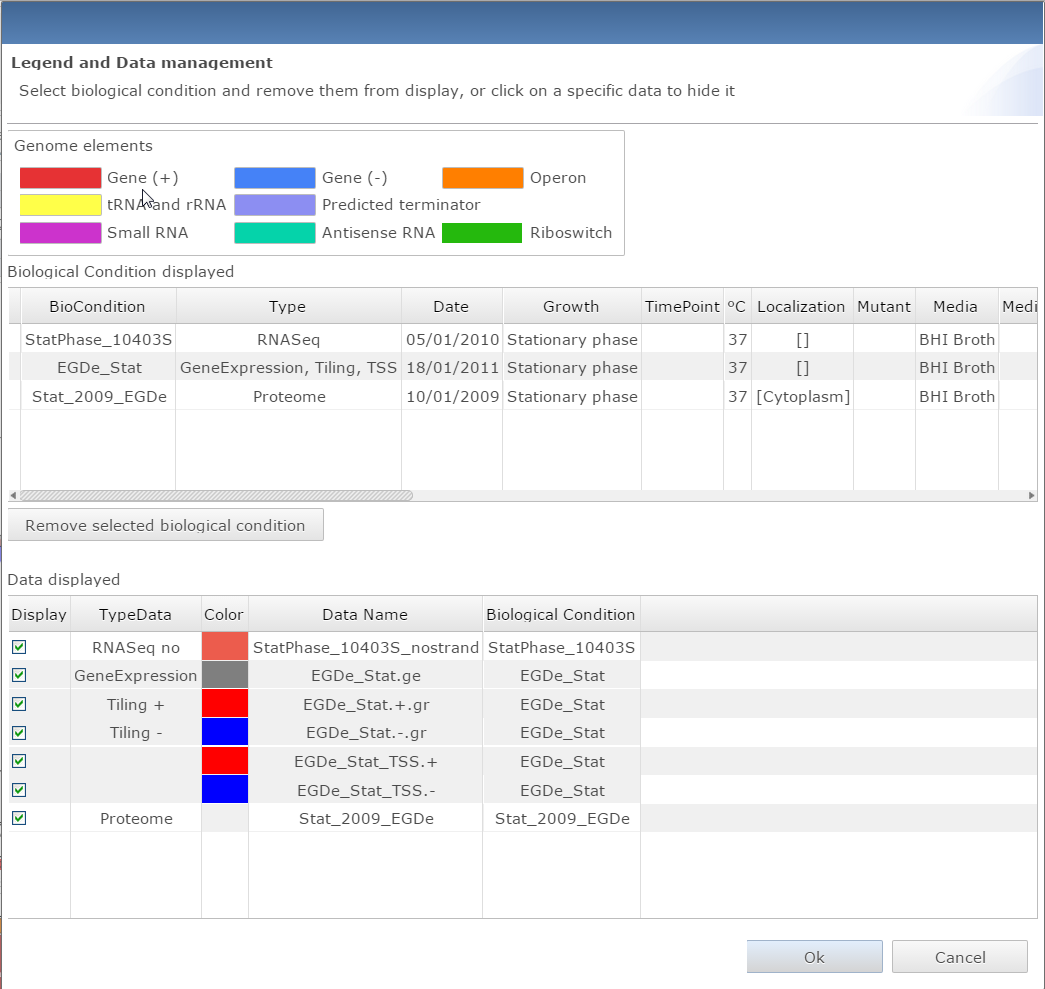 Dialog window for managin data et getting the legend for the genome viewer |
Manage data:
- Access the data legend and remove specific dataset from the view
- Save in text file the current genome viewer properties
- Change the display between absolute expression data (Log(expression)) to relative expression data (Log(Fold Change))
- Select one of three representation: Display by biological condition, by dataset, or overlap all dataset on one single track.
|
Omics data viewer

Omics data viewer of the genome viewer
 Omics data viewer of the genome viewer |
Click on Expression Atlas on the Omics selection panel
- RNASeq data: Select the value of Log(Fold Change) used to detect differential expression.
- Tiling array, Gene expression array and TSS data: Display selected datasets in the genome viewer in which the small RNA will be highlighted, or to the heatmap viewer to show the values of Log(Fold-change).
- Proteomic data: List of relative value expression data in which the small RNAis over-expressed (i.e. Log(FC) > cut-off).
- Move view vertically: List of relative value expression data in which the small RNAis under-expressed (i.e. Log(FC) < - cut-off).
|
Genome annotation

Genome annotation panel of the genome viewer
 Genome annotation panel of the genome viewer |
Click on Expression Atlas on the Omics selection panel
- Base pair condition: Select the value of Log(Fold Change) used to detect differential expression.
- Genome annotation: Display selected datasets in the genome viewer in which the small RNA will be highlighted.
- Move view horizontally: List of relative value expression data in which the small RNAis over-expressed (i.e. Log(FC) > cut-off).
|






