Transcriptomic tools
Contents
General organization
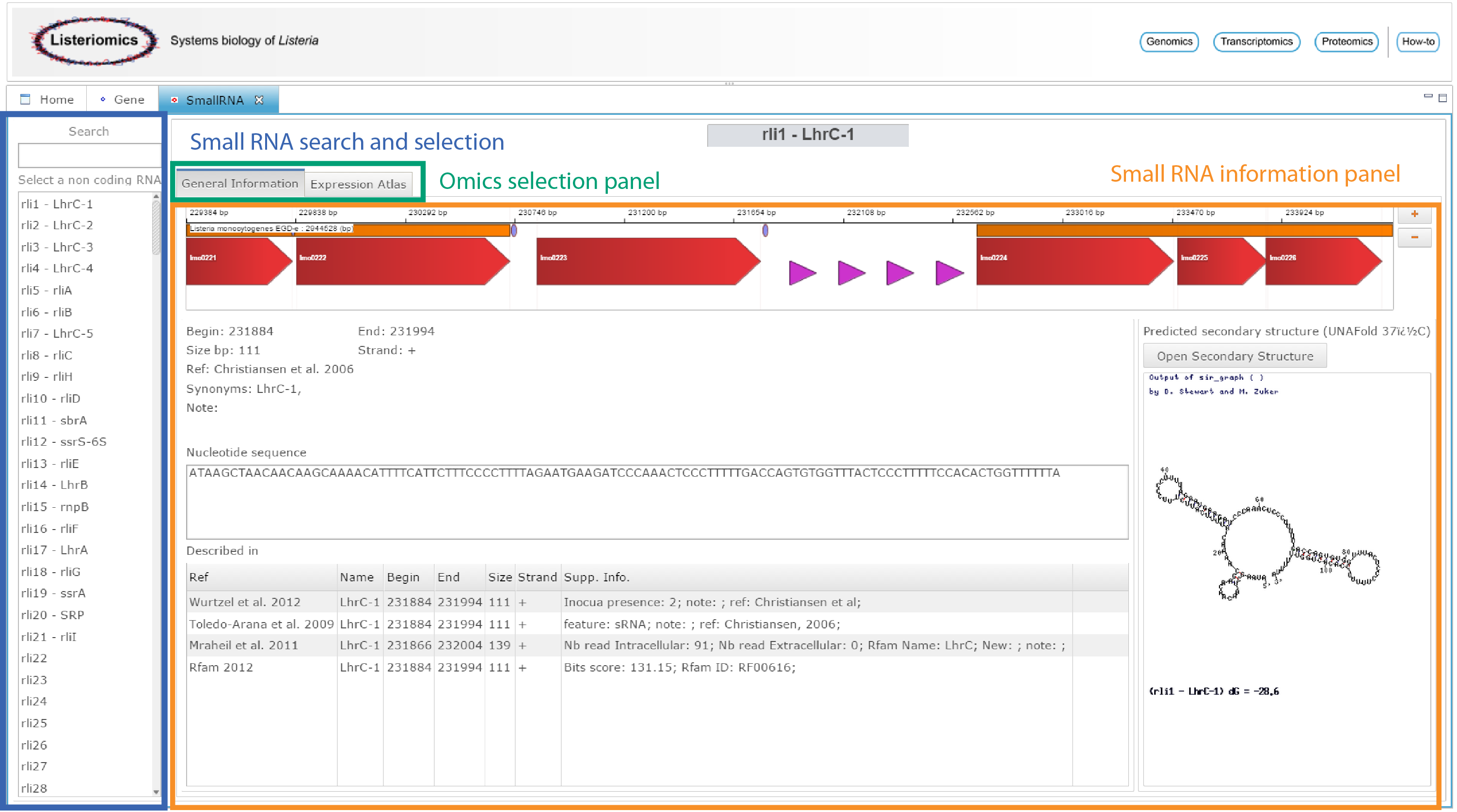
The small RNA webpage can be accessed either from the home webpage of the Listeriomics website or by double-clicking on a small RNA in the genome viewer.
| Small RNA search and selection
In this list, all the small RNA available in Listeria monocytogenes EGD-e strain are displayed. One can search for a small RNA. A click on an element of the list will display all related information on the Small RNA information panel. |
| Omics selection panel
For each Listeria small RNA two different information panels are available.
|
| Small RNA information panel
The panel in which all information will be displayed |
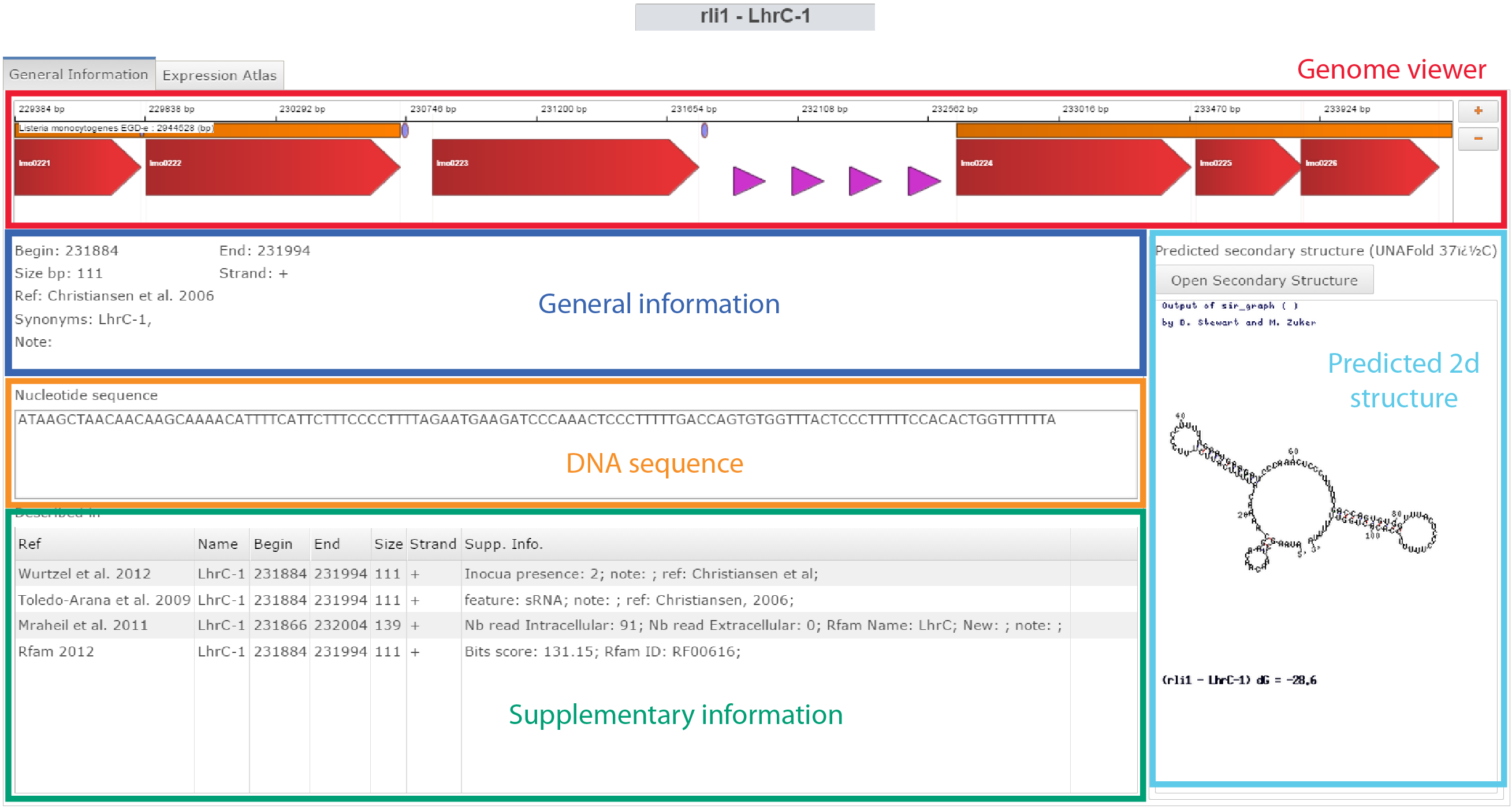
How-to access Small RNA information ?
Click on General information on the Omics selection panel
|
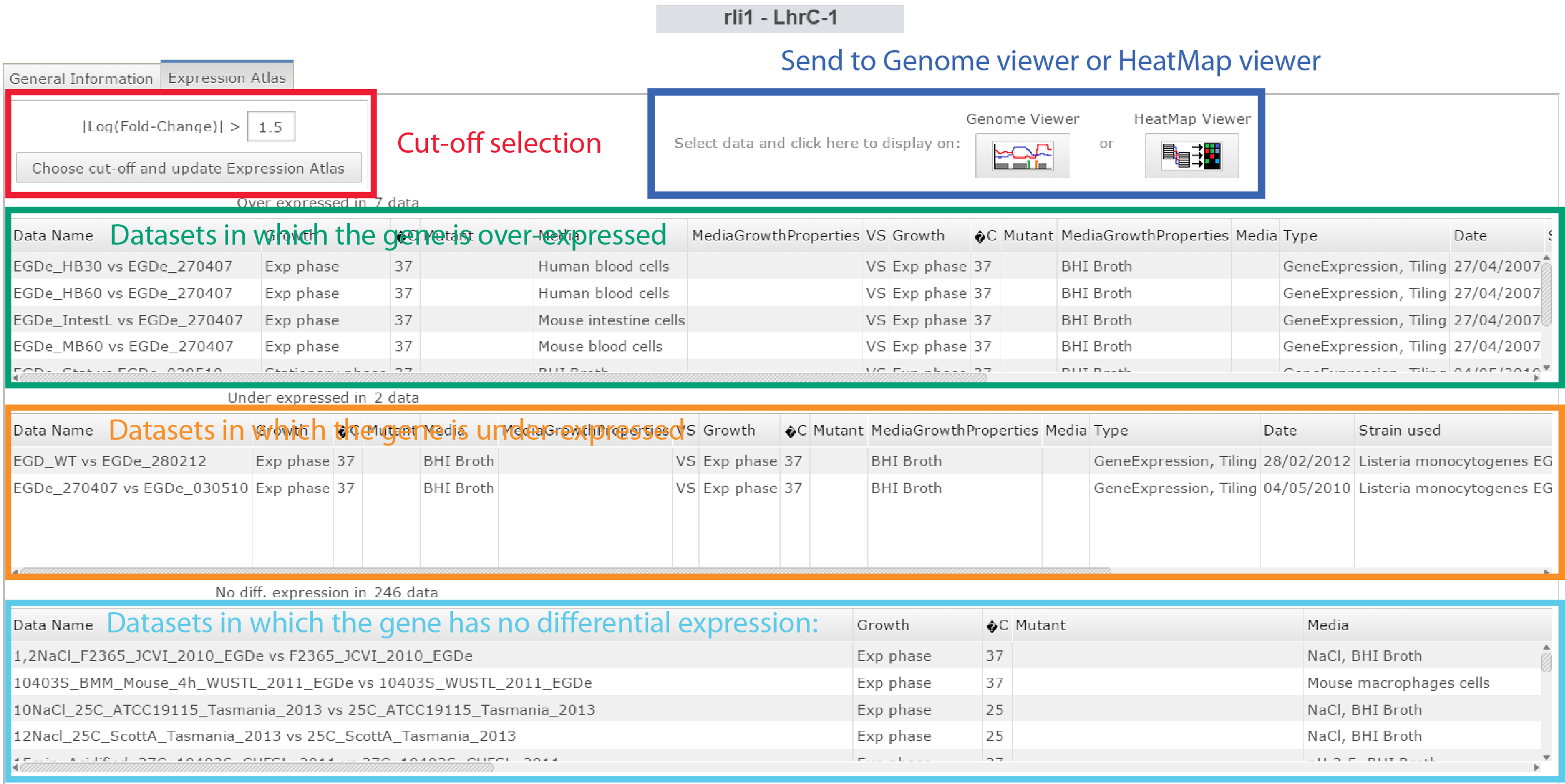
Is my Small RNA transcribed ?
Click on Expression Atlas on the Omics selection panel
|
Summary of Small RNA studies in Listeria monocytogenes EGD-e
In the past decade, noncoding RNAs in Listeria have been studied in detail
- 155 sRNAs (acting mainly in trans)
- 46 cis regulatory elements (cisRegs)
- 104 antisense RNAs (asRNAs)
Tables summarizing all the small RNAs observed can be accessed in the small RNAs summary webpage. This page can be accessed from the home webpage of the Listeriomics website.
| Small RNA type selection:
Type of small RNA to display:
|
| Small RNAs summary table
Table with different information on each small RNA:
By clicking on a small RNA, the small RNA webpage will open to give more information. |
Transcriptomic database
We downloaded the 64 Listeria related ArrayExpress (27) experiments in MAGE-TAB standard (36) (Table S3). Every MAGE-TAB file included an IDF (Investigation Description Format) file giving general information about the experiment, SDRF (Sample and Data Relationship Format) file giving data relationship, and processed data. We manually curated every SDRF file and integrated the different information about the data. We extracted the main biological attributes for each dataset and associated it to different key parameters: “TimePoint”, for growh time, void value being overnight growth; “Mutant” indicating a gene is removed or over-expressed; “Growth” indicating the growth phase from which the bacteria were extracted; “Temperature” of the growth media; “Media” properties for example if Glucose was added, or if bacteria were extracted from macrophages; “Strain used” and “Strain array” indicating the strain used for the experiment, and strain actually used for designing the array or mapping RNASeq reads. We then added matrices of expression or comparison for each biological condition. In total, 32 different gene expression array and 6 RNASeq technologies were combined. For Gene Expression arrays we downloaded the 32 ADF (Array Design Format) files which describe the relationship between array probes and genes. Only processed data provided by ArrayExpress were used, no new normalization was applied. Depending on the experiment, two types of data were available: Absolute expression value and relative expression value. The former if available, was log-transformed and median normalized, median differences of log values were calculated (LOGFC). For relative expression tables we were able to always extract LOGFC values. Fold change (FC) and t-test p-values were also extracted when available. For every type of data and value we calculated the mean value and standard deviation of each gene for technical and biological replicates. We did the same when many probes were available for one gene. In the case of RNASeq, raw reads alignments were performed with bowtie version 1.0.0 (parameters : --best –strata -k 1 -m 100 -n 1 -l 20) (37) or segemehl version 0.1.7-411 (parameters : default) (38) or novoalignCS (parameters : -F BFASTQ –s 1 –l 14 –t 50) Reads were mapped on L. mono. EGD-e genome sequence (NC_003210.1) for E-GEOD-47721, E-GEOD-51487, E-GEOD-60363, E-MTAB-329, PRJEB6949) and L. mono. 10403S (NC_017544) for E-GEOD-15651. Alignment files were then sorted and compressed to bam file format with SAMtools version 0.1.19 (39). BEDTools version 2.17.0 (40) was used in combination with custom python scripts to compute Reads Per Kilobase per Million mapped reads (RPKM) for each genomic feature and Reads Per Million (RPM) for genome-wide coverage. The differential expression analysis was performed using DESeq version 1.14.0 (41) following manual recommendation with respect to the number of available replicates on per feature raw counts.
A total of 492 files, 150 absolute value data, 342 relative expression data, (Figure 1) were created and integrated in the Listeriomics website. For every experiment, relative expression data were always available whereas absolute expression data was found only in a quarter of the data.
| ID | Accession | Title | Type | Organism | Assays | Year month / day | Processed Data | Raw Data |
|---|---|---|---|---|---|---|---|---|
| 1 | E-MEXP-1118 | Transcription profiling of Listeria monocytogenes EGD-e after heat-shock for 3, 10, 20, and 40 min | Listeria monocytogenes | 24 | 2007 10 / 9 | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-1118/E-MEXP-1118.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-1118/E-MEXP-1118.raw.1.zip | |
| 2 | E-BUGS-46 | Transcription profiling of Listeria monocytogenes relA and codY mutants | transcription profiling by array | Listeria monocytogenes | 6 | 2007 3 / 7 | http://www.ebi.ac.uk/arrayexpress/files/E-BUGS-46/E-BUGS-46.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-BUGS-46/E-BUGS-46.raw.1.zip |
| 3 | E-GEOD-12143 | Listeria monocytogenes EGD after growth BHI vs. LB vs. MM | transcription profiling by array | Listeria monocytogenes | 9 | 2008 11 / 30 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-12143/E-GEOD-12143.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-12143/E-GEOD-12143.raw.1.zip |
| 4 | E-GEOD-12145 | Listeria monocytogenes EGD?prfApPrfA and EGD?prfApPrfA* compared to the wild type strain EGD | transcription profiling by array | Listeria monocytogenes | 12 | 2008 11 / 30 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-12145/E-GEOD-12145.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-12145/E-GEOD-12145.raw.1.zip |
| 5 | E-GEOD-12146 | Listeria monocytogenes EGD and EGD-e | transcription profiling by array | Listeria monocytogenes, Listeria monocytogenes EGD-e | 3 | 2008 11 / 30 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-12146/E-GEOD-12146.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-12146/E-GEOD-12146.raw.1.zip |
| 6 | E-GEOD-12151 | Modulation of PrfA activity in Listeria monocytogenes upon growth in different culture media | transcription profiling by array | Listeria monocytogenes, Listeria monocytogenes EGD-e | 24 | 2008 11 / 30 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-12151/E-GEOD-12151.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-12151/E-GEOD-12151.raw.1.zip |
| 7 | E-GEOD-11459 | Glycerol metabolism and PrfA activity in Listeria monocytogenes | transcription profiling by array | Listeria monocytogenes | 23 | 2008 5 / 30 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-11459/E-GEOD-11459.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-11459/E-GEOD-11459.raw.1.zip |
| 8 | E-MEXP-1144 | Transcription profiling of wild type Listera monocytogenes EGD-e and an isogenic deltahlydeltaplcA mutant strain 1 hour or 4 hours post infection into human intestinal epithelial cells | transcription profiling by array | Listeria monocytogenes | 12 | 2008 6 / 18 | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-1144/E-MEXP-1144.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-1144/E-MEXP-1144.raw.1.zip |
| 9 | E-MEXP-1162 | Transcription profiling of Listeria monocytogenes EGD-e versus the over-expressing strain (EGD-e+pPXrnhB | transcription profiling by array | Listeria monocytogenes | 6 | 2008 6 / 29 | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-1162/E-MEXP-1162.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-1162/E-MEXP-1162.raw.1.zip |
| 10 | E-MEXP-1170 | Transcription profiling of wild type and sigma B mutant Liseteria monocytogenes to characterise the EGD-e sigma B regulon | transcription profiling by array | Listeria monocytogenes | 26 | 2008 7 / 2 | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-1170/E-MEXP-1170.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-1170/E-MEXP-1170.raw.1.zip |
| 11 | E-MEXP-2138 | Transcription profiling of Listeria monocytogenes wild type and prfA, sigB and hfq deletion mutants grown in different media and at different temperatures | transcription profiling by array | Listeria monocytogenes | 39 | 2009 4 / 24 | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-2138/E-MEXP-2138.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-2138/E-MEXP-2138.raw.1.zip |
| 12 | E-MEXP-2142 | Tiling path analysis of Listeria monocytogenes wild type and prfA, sigB and hfq deletion mutants grown in different media and at different temperatures | transcription profiling by tiling array | Listeria monocytogenes | 42 | 2009 4 / 24 | Data is not available | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-2142/E-MEXP-2142.raw.1.zip |
| 13 | E-GEOD-19918 | Transcriptomic and phenotypic responses of L. monocytogenes with different growth efficiencies under acidic conditions | transcription profiling by array | Listeria monocytogenes | 14 | 2010 1 / 15 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-19918/E-GEOD-19918.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-19918/E-GEOD-19918.raw.1.zip |
| 14 | E-GEOD-15651 | Listeria monocytogenes Transcriptome with RNA-Seq | RNA-seq of coding RNA | Listeria monocytogenes | 4 | 2010 1 / 5 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-15651/processed/ | Data is not available |
| 15 | E-GEOD-24107 | The cell envelope stress response mediated by the LiaFSRLm three-component system of Listeria monocytogenes is controlled via the phosphatase activity of the bifunctional histidine kinase LiaSLm | transcription profiling by array | Listeria monocytogenes EGD-e | 11 | 2010 11 / 16 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-24107/E-GEOD-24107.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-24107/E-GEOD-24107.raw.1.zip |
| 16 | E-MEXP-1947 | Transcription profiling of Listeria monocytogenes after infection into murine macrophages | transcription profiling by array | Listeria monocytogenes | 55 | 2010 11 / 25 | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-1947/E-MEXP-1947.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-1947/E-MEXP-1947.raw.1.zip |
| 17 | E-GEOD-16336 | Induction of virulence genes in Listeria monocytogenes serotype 4b upon exposure to concentrations of biocides | transcription profiling by tiling array | Listeria monocytogenes str. 4b F2365 | 34 | 2010 11 / 27 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-16336/E-GEOD-16336.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-16336/E-GEOD-16336.raw.1.zip |
| 18 | E-GEOD-19014 | Pyruvate carboxylase and carbon metabolism in Listeria monocytogenes | transcription profiling by array | Listeria monocytogenes | 4 | 2010 2 / 9 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-19014/E-GEOD-19014.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-19014/E-GEOD-19014.raw.1.zip |
| 19 | E-TABM-663 | Transcription profiling of Listeria monocytogenes intracellularly replicating in mouse macrophages activated by IFN-gamma | transcription profiling by array | Listeria monocytogenes | 12 | 2010 3 / 11 | http://www.ebi.ac.uk/arrayexpress/files/E-TABM-663/E-TABM-663.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-TABM-663/E-TABM-663.raw.1.zip |
| 20 | E-GEOD-20367 | Probing the pan genome of a foodborne bacterial pathogen Listeria monocytogenes | comparative genomic hybridization by array | Listeria monocytogenes, Listeria monocytogenes EGD-e, Listeria monocytogenes FSL F2-208, Listeria monocytogenes FSL J1-208, Listeria monocytogenes FSL J2-071 | 18 | 2010 4 / 6 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-20367/E-GEOD-20367.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-20367/raw/ |
| 21 | E-MTAB-116 | Transcription profiling of wild type and L forms of Listeria monocytogenes | transcription profiling by array | Listeria monocytogenes | 4 | 2010 5 / 12 | Data is not available | http://www.ebi.ac.uk/arrayexpress/files/E-MTAB-116/E-MTAB-116.raw.1.zip |
| 22 | E-GEOD-20274 | Transcriptome profiling of Listeria monocytogenes during growth on a RTE-meat matrix | transcription profiling by array | Listeria monocytogenes | 12 | 2010 5 / 14 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-20274/E-GEOD-20274.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-20274/E-GEOD-20274.raw.1.zip |
| 23 | E-GEOD-19570 | Osmotic stress in Listeria monocytogenes serotype 4b strain F2365 grown in BHI medium and BHI medium with 1.2% NaCl | transcription profiling by array | Listeria monocytogenes | 12 | 2010 5 / 15 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-19570/E-GEOD-19570.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-19570/E-GEOD-19570.raw.1.zip |
| 24 | E-GEOD-11347 | Characterization of the SigB-PrfA regulatory network in Listeria monocytogenes | transcription profiling by array | Listeria monocytogenes | 24 | 2010 5 / 19 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-11347/E-GEOD-11347.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-11347/E-GEOD-11347.raw.1.zip |
| 25 | E-GEOD-12634 | Listeria monocytogenes SOS response determined after mitomycin C exposure | transcription profiling by array | Listeria monocytogenes EGD-e | 16 | 2010 5 / 19 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-12634/E-GEOD-12634.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-12634/E-GEOD-12634.raw.1.zip |
| 26 | E-GEOD-18363 | Identification of the major PEP-phosphotransferase systems (PTS) for glucose, mannose and cellobiose of L. monocytogenes | transcription profiling by array | Listeria monocytogenes EGD-e | 24 | 2010 5 / 19 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-18363/E-GEOD-18363.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-18363/E-GEOD-18363.raw.1.zip |
| 27 | E-GEOD-21427 | Sigma B in Listeria monocytogenes strains from different lineages | transcription profiling by array | Listeria monocytogenes | 4 | 2010 6 / 1 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-21427/E-GEOD-21427.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-21427/E-GEOD-21427.raw.1.zip |
| 28 | E-GEOD-6421 | regulation of Listeria monocytogenes internalin-like genes by ?B and PrfA as revealed by subgenomic microarray analyses | transcription profiling by array | Listeria monocytogenes, Listeria monocytogenes 10403S | 4 | 2010 6 / 11 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-6421/E-GEOD-6421.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-6421/E-GEOD-6421.raw.1.zip |
| 29 | E-GEOD-18796 | Physiology and genetic activity of Listeria monocytogenes during multi-hurdle stress induced cell death | transcription profiling by array | Listeria monocytogenes | 8 | 2010 6 / 20 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-18796/E-GEOD-18796.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-18796/E-GEOD-18796.raw.1.zip |
| 30 | E-MTAB-118 | Transcription profiling of wild type and SreA mutant Listeria monocytogenes | transcription profiling by array | Listeria monocytogenes | 4 | 2010 6 / 20 | Data is not available | http://www.ebi.ac.uk/arrayexpress/files/E-MTAB-118/E-MTAB-118.raw.1.zip |
| 31 | E-GEOD-13057 | In vivo transcriptome of Listeria monocytogenes EGDe | transcription profiling by array | Listeria monocytogenes EGD-e | 8 | 2010 6 / 25 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-13057/E-GEOD-13057.processed.1.zip | Data is not available |
| 32 | E-MEXP-2279 | Transcription profiling of Listeria monocytogenes following oxygen restriction | transcription profiling by array | Listeria monocytogenes | 3 | 2010 7 / 1 | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-2279/processed/ | http://www.ebi.ac.uk/arrayexpress/files/E-MEXP-2279/E-MEXP-2279.raw.1.zip |
| 33 | E-GEOD-25195 | Transcriptional response of Listeria monocytogenes h7858 and f6854 to lactate and acetate | transcription profiling by array | Listeria monocytogenes | 32 | 2011 11 / 3 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-25195/E-GEOD-25195.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-25195/E-GEOD-25195.raw.1.zip |
| 34 | E-GEOD-34083 | Intracellular transcription by multiple Listeria monocytogenes strains | transcription profiling by array | Listeria monocytogenes | 4 | 2011 12 / 2 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-34083/E-GEOD-34083.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-34083/E-GEOD-34083.raw.1.zip |
| 35 | E-GEOD-16887 | Identification of sigB inhibitors in L. monocytogenes | transcription profiling by array | Listeria monocytogenes 10403S | 1 | 2011 12 / 6 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-16887/E-GEOD-16887.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-16887/E-GEOD-16887.raw.1.zip |
| 36 | E-GEOD-26690 | Transcriptomic Response of Listeria monocytogenes throughout the Life Cycle | transcription profiling by array | Listeria monocytogenes serotype 4b str. F2365 | 12 | 2011 2 / 24 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-26690/E-GEOD-26690.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-26690/E-GEOD-26690.raw.1.zip |
| 37 | E-GEOD-28507 | Listeria monocytogenes transcriptional response to bile | transcription profiling by array | Listeria monocytogenes | 6 | 2011 4 / 12 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-28507/E-GEOD-28507.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-28507/E-GEOD-28507.raw.1.zip |
| 38 | E-BUGS-110 | Global Transcriptional Analysis of Spontaneous Sakacin P-Resistant Mutant Strains of Listeria monocytogenes during Growth on Different Sugars | transcription profiling by array | Listeria monocytogenes | 16 | 2011 4 / 15 | http://www.ebi.ac.uk/arrayexpress/files/E-BUGS-110/processed/ | http://www.ebi.ac.uk/arrayexpress/files/E-BUGS-110/E-BUGS-110.raw.1.zip |
| 39 | E-GEOD-22819 | Gene expression profiling of a pressure-tolerant Listeria monocytogenes Scott A ctsR deletion mutant | transcription profiling by array | Listeria monocytogenes | 4 | 2011 6 / 10 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-22819/E-GEOD-22819.processed.1.zip | Data is not available |
| 40 | E-GEOD-22672 | Listeria monocytogenes Grown at Refrigeration Temperature Shows Reduced Acid Survival and an Altered Transcriptional Response to Acid Shock | transcription profiling by array | Listeria monocytogenes | 10 | 2011 6 / 30 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-22672/E-GEOD-22672.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-22672/E-GEOD-22672.raw.1.zip |
| 41 | E-GEOD-27521 | Temporal changes in Listeria monocytogenes gene expression during salt stress | transcription profiling by array | Listeria monocytogenes | 60 | 2011 7 / 1 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-27521/E-GEOD-27521.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-27521/E-GEOD-27521.raw.1.zip |
| 42 | E-MTAB-329 | non-coding RNA-Seq of Listeria monocytogenes from bacteria growing extracellularly and in macrophages | Listeria monocytogenes EGD-e | 2 | 2011 7 / 21 | http://www.ebi.ac.uk/arrayexpress/files/E-MTAB-329/E-MTAB-329.processed.1.zip | Data is not available | |
| 43 | E-GEOD-32434 | Analysis of ?L dependent traits based on phenotypic and transcriptomic evaluation of the Listeria monocytogenes EGDe strain | transcription profiling by array | Listeria monocytogenes EGD-e | 36 | 2012 1 / 1 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-32434/E-GEOD-32434.processed.1.zip | Data is not available |
| 44 | E-GEOD-27936 | Gene Expression Profiling of Listeria monocytogenes in monoculture and in co-culture state in the presence of Bacillus subtilis both as planktonic cells and in biofilms. | transcription profiling by array | Listeria monocytogenes J0161 | 8 | 2012 1 / 15 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-27936/E-GEOD-27936.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-27936/E-GEOD-27936.raw.1.zip |
| 45 | E-GEOD-32913 | Genome-wide transcriptional profiling of the cell envelope stress response of Listeria monocytogenes | transcription profiling by array | Listeria monocytogenes | 16 | 2012 1 / 24 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-32913/E-GEOD-32913.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-32913/E-GEOD-32913.raw.1.zip |
| 46 | E-GEOD-40598 | Role of a GntR-family response regulator LbrA in Listeria monocytogenes biofilm formation | transcription profiling by array | Listeria monocytogenes | 6 | 2012 9 / 6 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-40598/E-GEOD-40598.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-40598/E-GEOD-40598.raw.1.zip |
| 47 | E-GEOD-32172 | L. monocytogenes ctsR mutant 2-1 cells response to pressure | transcription profiling by array | Listeria monocytogenes str. Scott A | 2 | 2013 2 / 14 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-32172/E-GEOD-32172.processed.1.zip | Data is not available |
| 48 | E-GEOD-41891 | Gene expression profiling of a nisin-sensitive Listeria monocytogenes Scott A ctsR deletion mutant | transcription profiling by array | Listeria monocytogenes str. Scott A | 4 | 2013 2 / 14 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-41891/E-GEOD-41891.processed.1.zip | Data is not available |
| 49 | E-GEOD-46182 | Characterization of the lag-phase associated cold acclimation gene expression responses in L. monocytogenes EGD-e | transcription profiling by array | Listeria monocytogenes EGD-e | 12 | 2013 4 / 19 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-46182/E-GEOD-46182.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-46182/E-GEOD-46182.raw.1.zip |
| 50 | E-GEOD-43052 | The whole genome transcription analysis reveals the regulatory network for Listeria monocytogenes biofilm formation is modified in the prfA deletion mutant | transcription profiling by array | Listeria monocytogenes EGD-e | 15 | 2013 5 / 1 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-43052/E-GEOD-43052.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-43052/E-GEOD-43052.raw.1.zip |
| 51 | E-GEOD-46612 | Characterization of the transcriptomes of genetically diverse Listeria monocytogenes exposed to hyperosmotic and low temperature conditions | transcription profiling by array | Listeria monocytogenes | 6 | 2013 5 / 4 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-46612/E-GEOD-46612.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-46612/E-GEOD-46612.raw.1.zip |
| 52 | E-GEOD-42730 | Listeria monocytogenes strain F2365 has reduced ability to survive stress while maintaining virulence characteristics | transcription profiling by array | Listeria monocytogenes | 1 | 2013 6 / 1 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-42730/E-GEOD-42730.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-42730/E-GEOD-42730.raw.1.zip |
| 53 | E-GEOD-47721 | Transcriptomic profiling of L-rhamnose utilization in Lmo0753 deletion mutant of Listeria monocytogenes EGDe from human foodborne outbreak associated lineage LII | RNA-seq of coding RNA | Listeria monocytogenes | 4 | 2013 17 / 7 | Data is not available | Data is not available |
| 54 | E-GEOD-49056 | Listeria monocytogenes Sigma B, but not PrfA, shows strain specific contributions to growth under salt stress, and prolongated lag phase of a sigB null mutant across strains | transcription profiling by array | Listeria monocytogenes | 1 | 2013 20 / 7 | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-49056/E-GEOD-49056.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-GEOD-49056/E-GEOD-49056.raw.1.zip |
| 55 | E-MTAB-1677 | Transcription profiling by tiling array of Listeria monocytogenes EGD wild type, Listeria monocytogenes EGD-e wild-type and EGD-e PrfA* | transcription profiling by tiling array | Listeria monocytogenes EGD, Listeria monocytogenes EGD-e | 9 | 2013 8 / 1 | http://www.ebi.ac.uk/arrayexpress/files/E-MTAB-1677/E-MTAB-1677.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-MTAB-1677/E-MTAB-1677.raw.1.zip |
| 56 | E-MTAB-1676 | Transcription profiling by array of Listeria monocytogenes EGD wild type, Listeria monocytogenes EGD-e wild-type and EGD-e PrfA* | transcription profiling by array | Listeria monocytogenes EGD, Listeria monocytogenes EGD-e | 9 | 2013 8 / 1 | http://www.ebi.ac.uk/arrayexpress/files/E-MTAB-1676/E-MTAB-1676.processed.1.zip | http://www.ebi.ac.uk/arrayexpress/files/E-MTAB-1676/E-MTAB-1676.raw.1.zip |
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedWurtzel