Gene webpage
The gene webpage can be accessed either from the home webpage of the Listeriomics website or by double-clicking on a genome in the genomic webpage.
General organization
| Chromosome selection
Here one can select a specific genome among the 50 Listeria strains available. Then one has to select a chromosome. In Listeria species, in general the first one is a circular chromosome and the second one is a plasmid. With a star are indicated genomes for which transcriptomic datasets are available.
|
| Gene search and selection
In this list, all the genes available for a specific chromosome are displayed. One can search for a gene locus, a gene name, or even key words which can be found in the annotation of each gene. A click on an element of the list will display all related information on the Gene information panel.
|
| Omics selection panel
For each Listeria gene four different information panels are available.
- General information: The position of the gene, possible annotated function, nucleotide sequence and amino-acid sequence.
- Gene conservation in other Listeria strains : It dynamically displays on the Listeria phylogram presence of absence of homolog to the selected gene in each existing Listeria strain. It also displays a summary table of all homologs with their similarity percentages and amino acid sequences. One can then create a multi-alignment file of the homologs.
- Expression atlas: It shows in which transcriptomics datasets the selected gene is differently expressed. The cut-off on the Log(Fold Change) value for detecting such elements can be changed by the user.
- Protein atlas: It displays every proteomics dataset in which the protein coded by the selected gene has been detected.
|
| Gene information panel
The panel in which all information will be displayed
|
How-to access gene annotation ?
|
|
Click on General information on the Omics selection panel
- Genome viewer: Displays the gene in the genome viewer to see surrounding genome features.
- General information: Every general information available on gene position, product, Cluster of Orthologous Gene (COG) and summary of gene conservation.
- Supplementary information: All supplementary information available on the gene annotation such as conserved motif or alternative annotation.
- Sequences: DNA and amino acid sequences are available here for copy/paste.
|
Is my gene conserved ?
 Gene conservation panel of the Gene webpage |
Click on General information on the Omics selection panel
- Genome viewer: Displays the gene in the genome viewer to see surrounding genome features.
- General information: Every general information available on gene position, product, Cluster of Orthologous Gene (COG) and summary of gene conservation.
- Supplementary information: All supplementary information available on the gene annotation such as conserved motif or alternative annotation.
- Sequences: DNA and amino acid sequences are available here for copy/paste.
|
Is my gene transcribed ?
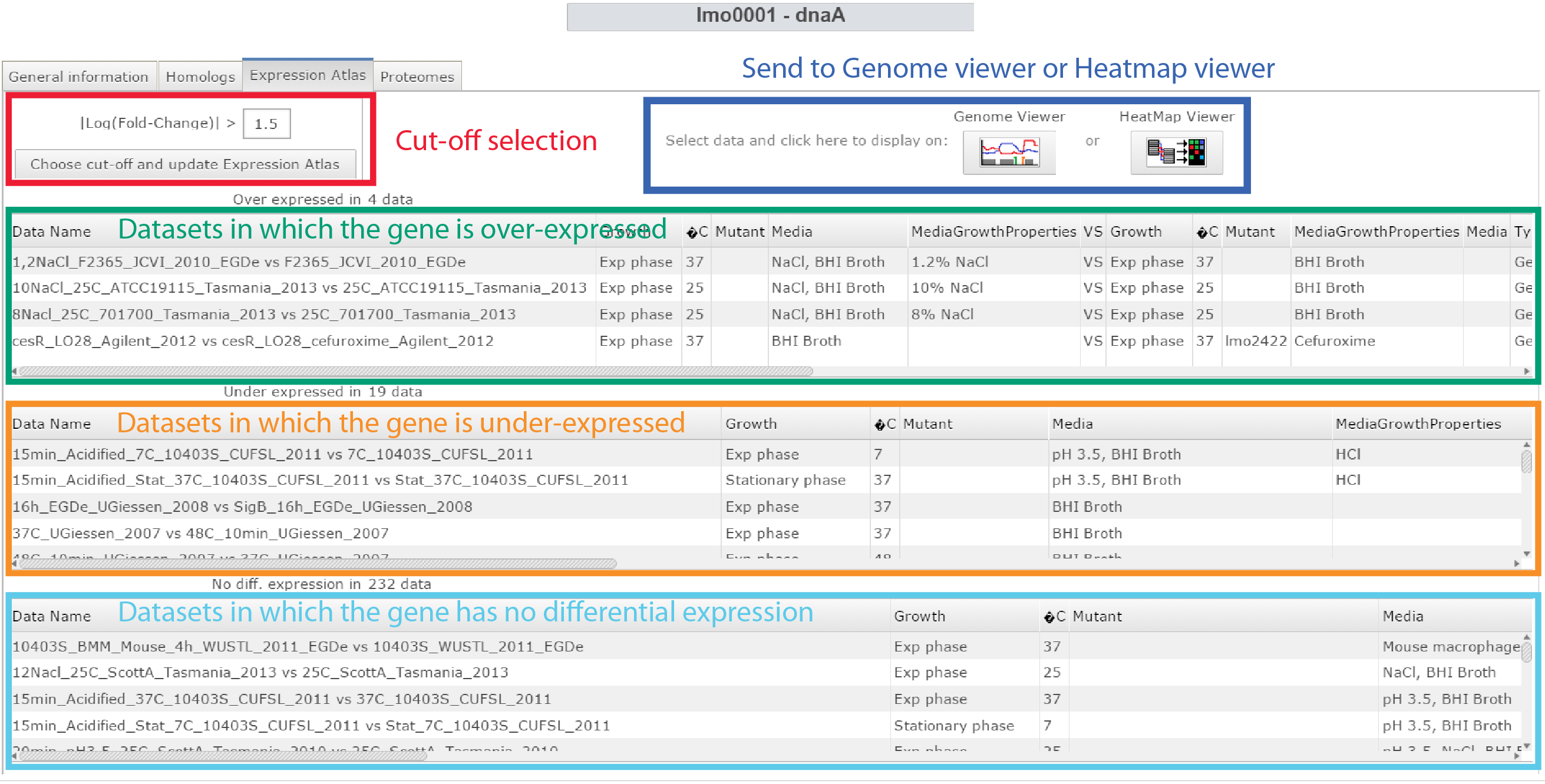 Expression atlas panel of the Gene webpage |
Click on General information on the Omics selection panel
- Genome viewer: Displays the gene in the genome viewer to see surrounding genome features.
- General information: Every general information available on gene position, product, Cluster of Orthologous Gene (COG) and summary of gene conservation.
- Supplementary information: All supplementary information available on the gene annotation such as conserved motif or alternative annotation.
- Sequences: DNA and amino acid sequences are available here for copy/paste.
|
Is my gene translated ?
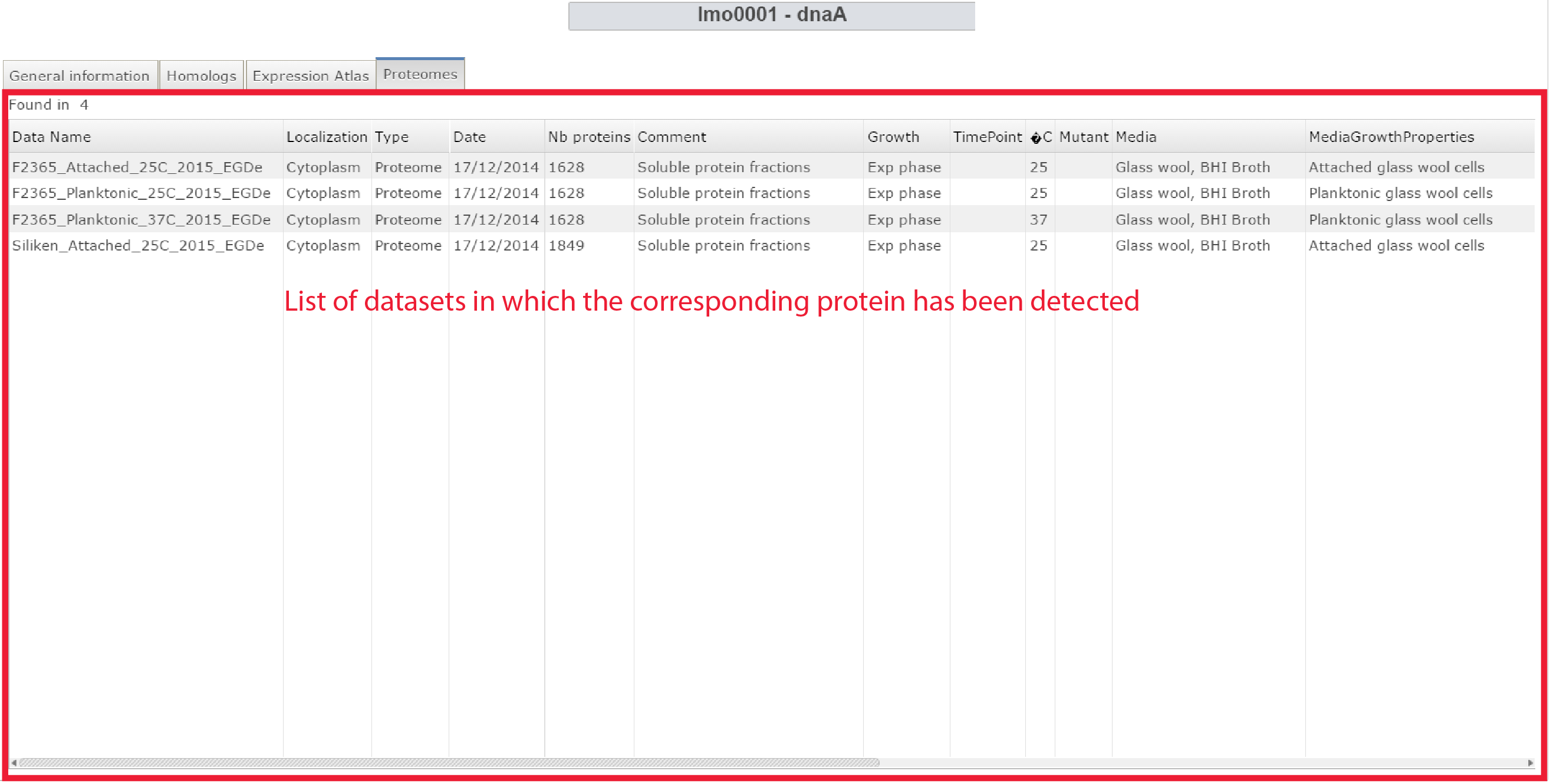 Protein atlas panel of the Gene webpage |
Click on General information on the Omics selection panel
- Genome viewer: Displays the gene in the genome viewer to see surrounding genome features.
- General information: Every general information available on gene position, product, Cluster of Orthologous Gene (COG) and summary of gene conservation.
- Supplementary information: All supplementary information available on the gene annotation such as conserved motif or alternative annotation.
- Sequences: DNA and amino acid sequences are available here for copy/paste.
|


