Difference between revisions of "Summary"
(→Screenshots) |
(Undo revision 147960 by 157.99.69.252 (talk)) |
||
| (11 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[File:HomePageListeriomics.png|thumb|300px|right|Home page of the Listeriomics website]] | [[File:HomePageListeriomics.png|thumb|300px|right|Home page of the Listeriomics website]] | ||
| − | Listeriomics integrates all complete genomes, transcriptomes and proteomes published for <i>Listeria</i> species to date. It allows navigating among all these datasets with enriched metadata in a user-friendly format. | + | Listeriomics integrates all complete genomes, transcriptomes and proteomes published for <i>Listeria</i> species to date. It allows navigating among all these datasets with enriched metadata in a user-friendly format. |
The main purpose of the website is to give scientists a quick and easy access to tools created for answering the four questions one has when starting a study on a specific genomic element: | The main purpose of the website is to give scientists a quick and easy access to tools created for answering the four questions one has when starting a study on a specific genomic element: | ||
| Line 7: | Line 7: | ||
# What are the biological conditions in which a genomic feature is translated? | # What are the biological conditions in which a genomic feature is translated? | ||
# What is the regulation network involved with a genomic feature of interest? | # What is the regulation network involved with a genomic feature of interest? | ||
| − | |||
| − | |||
| − | |||
| − | |||
The website integrates different type of tools for “omics” data analyses: | The website integrates different type of tools for “omics” data analyses: | ||
| Line 17: | Line 13: | ||
* A tool to explore gene conservation throughout ''Listeria'' strains. | * A tool to explore gene conservation throughout ''Listeria'' strains. | ||
* A co-expression network tool to discover new potential regulation. | * A co-expression network tool to discover new potential regulation. | ||
| + | |||
| + | |||
= Tutorials = | = Tutorials = | ||
| Line 25: | Line 23: | ||
| General organization of the website and principal features | | General organization of the website and principal features | ||
|} | |} | ||
| + | |||
| + | === Tools === | ||
| + | |||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| Line 41: | Line 42: | ||
| [[Proteomic tools]] | | [[Proteomic tools]] | ||
| How-to access proteomic dataset and display it or perform an analysis with the protein atlas | | How-to access proteomic dataset and display it or perform an analysis with the protein atlas | ||
| − | + | |- | |
| + | | [[Co-Expression_network_tools|Co-expression network tools]] | ||
| + | | How-to access to the co-expression network and search for potential co-regulation of genomic feature | ||
| + | |- | ||
| + | | [[Submit_Data|Submit your datasets]] | ||
| + | | How-to access to the submit section of Listeriomics in which you can inform us of important database update to make | ||
| + | |} | ||
| + | |||
| + | === Viewers === | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | | [[Genome viewer]] | ||
| + | | How-to use the genome viewer for visualizing "omics" datasets | ||
| + | |- | ||
| + | | [[Transcriptomic HeatMap viewer]] | ||
| + | | How-to use the transcriptomic heatmap viewer and decipher differently expressed genomic features | ||
| + | |- | ||
| + | | [[Proteomic HeatMap viewer]] | ||
| + | | How-to use the proteomic heatmap viewer and decipher detected protein in every proteomic experiment | ||
|} | |} | ||
| Line 53: | Line 73: | ||
* 74 proteome datasets | * 74 proteome datasets | ||
| + | The whole database can be downloaded [http://listeriomics01.hosting.pasteur.fr/DatabaseListeriomics.zip Here] | ||
= Screenshots = | = Screenshots = | ||
<gallery widths=200px> | <gallery widths=200px> | ||
Image:BHIViewer.png|Genome viewer | Image:BHIViewer.png|Genome viewer | ||
| + | Image:GenomeviewerChangeView.png|Example of the zoom capability of the Genome Viewer | ||
Image:genomes.png|Genome webpage | Image:genomes.png|Genome webpage | ||
Image:Genepanel.png|Gene webpage | Image:Genepanel.png|Gene webpage | ||
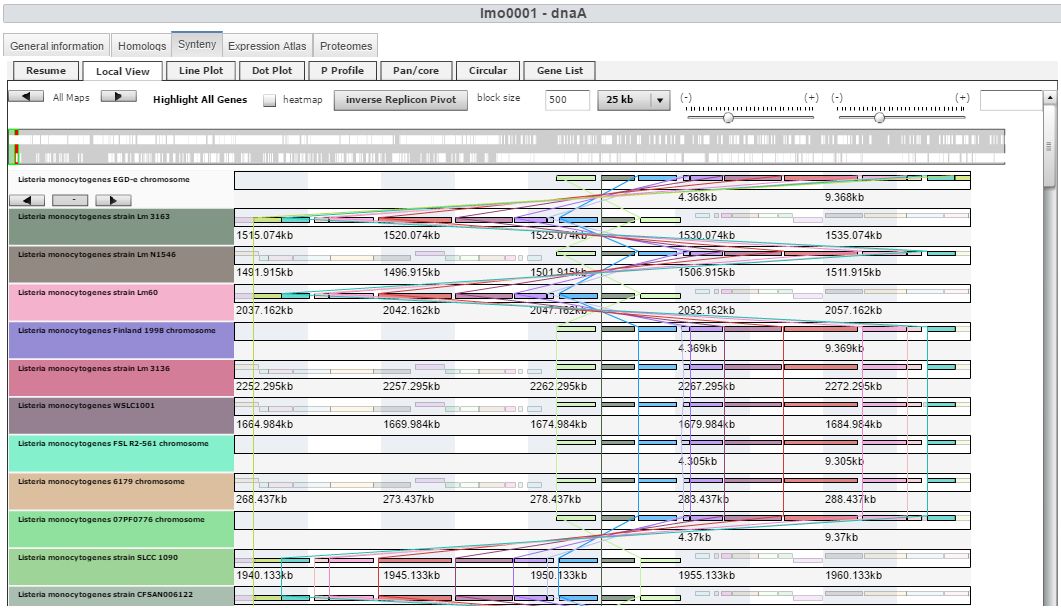
| + | Image:Synteny.png|Synteny webpage | ||
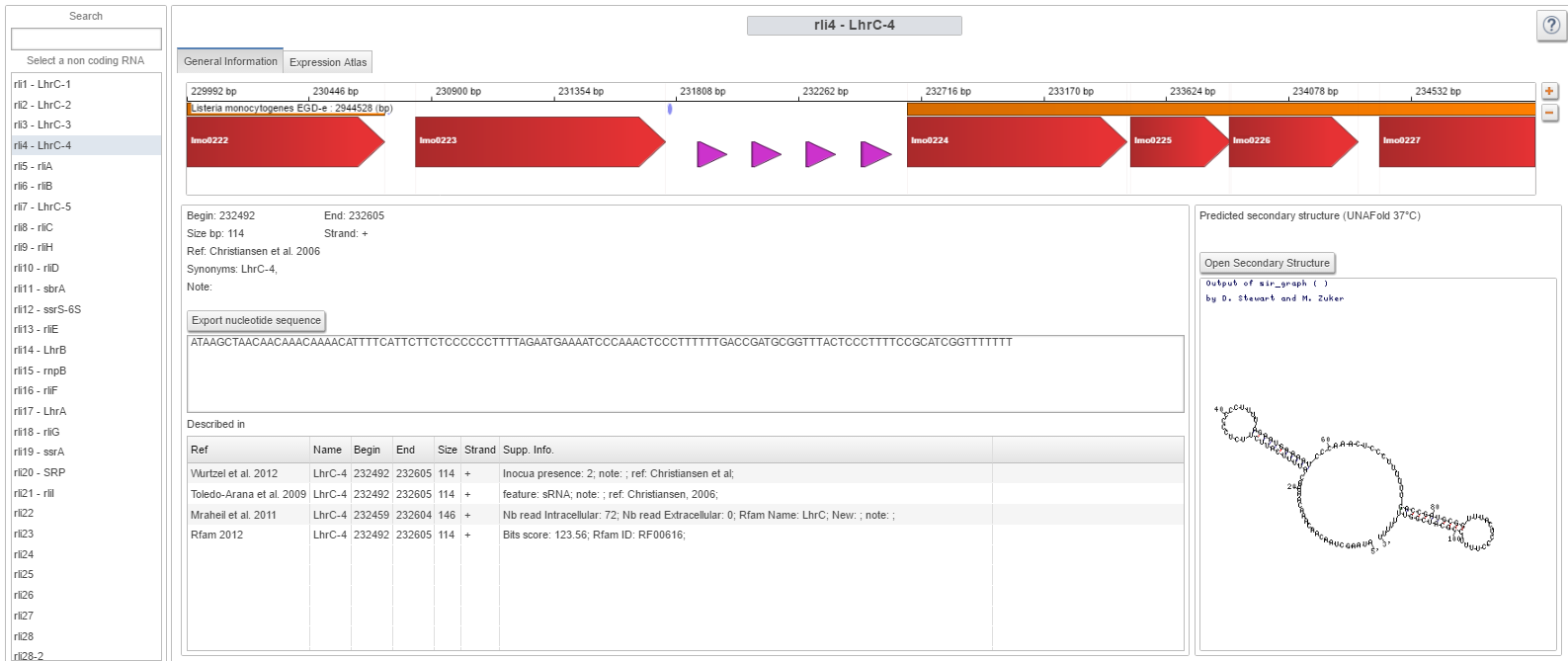
Image:smallRNA.png|Small RNA webpage | Image:smallRNA.png|Small RNA webpage | ||
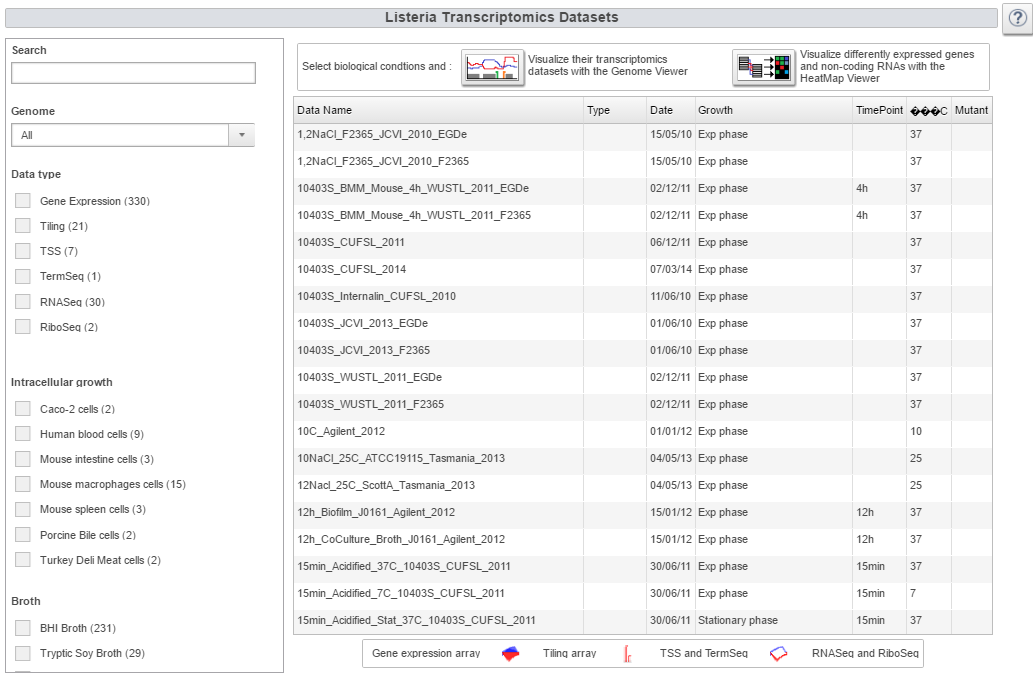
Image:transcriptome.png|Transcriptome webpage | Image:transcriptome.png|Transcriptome webpage | ||
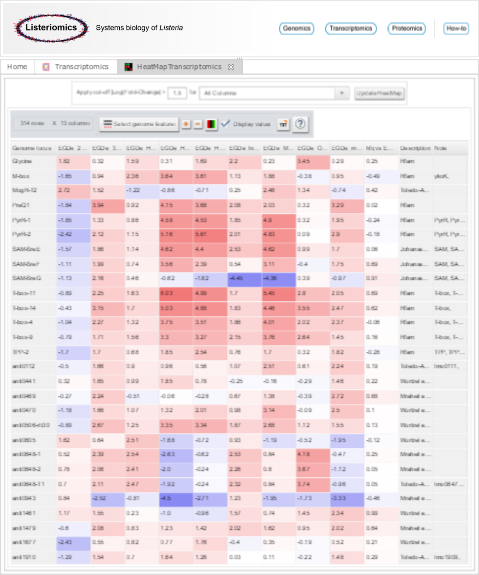
| + | Image:Heatmap.png|Heatmap visualization of transcriptomics datasets | ||
Image:proteome.png|Proteome webpage | Image:proteome.png|Proteome webpage | ||
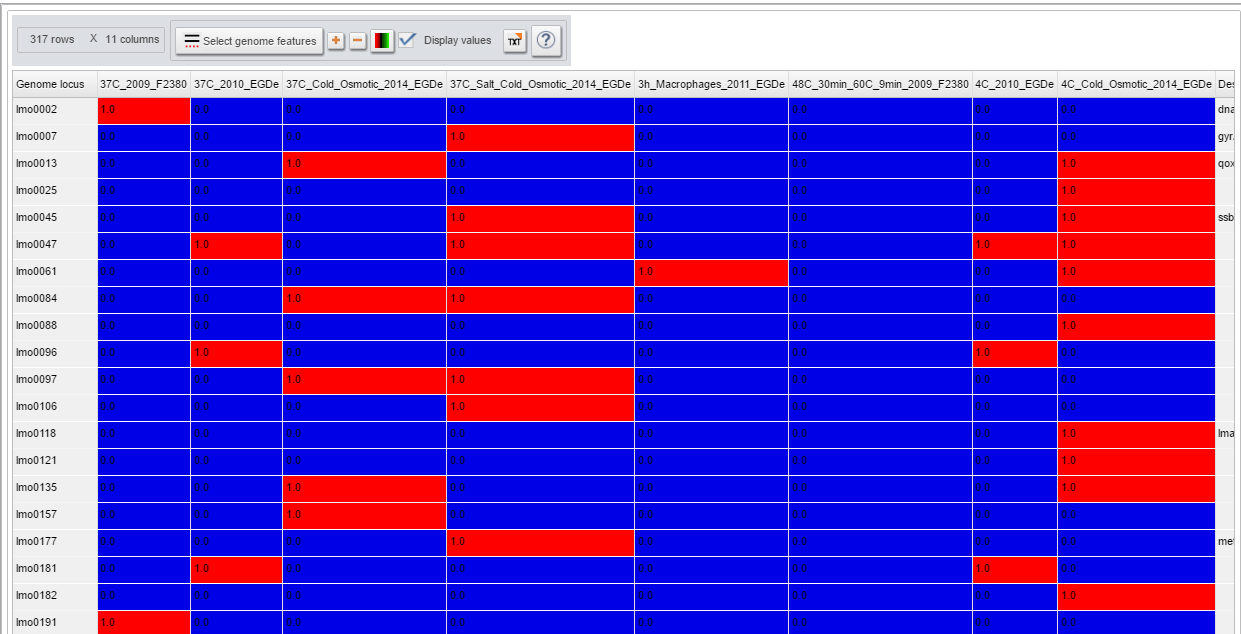
| + | Image:HeatmapProteomic.png|Heatmap visualization of proteomics datasets | ||
Image:SeqInfo.png|Sequence information panel | Image:SeqInfo.png|Sequence information panel | ||
Image:coexpression.png|Co-expression network webpage | Image:coexpression.png|Co-expression network webpage | ||
</gallery> | </gallery> | ||
Latest revision as of 14:01, 29 November 2022
Listeriomics integrates all complete genomes, transcriptomes and proteomes published for Listeria species to date. It allows navigating among all these datasets with enriched metadata in a user-friendly format.
The main purpose of the website is to give scientists a quick and easy access to tools created for answering the four questions one has when starting a study on a specific genomic element:
- What are the known functions of a genomic feature in a given strain and homologies in closely related strains?
- What are the biological conditions in which a genomic feature is transcribed?
- What are the biological conditions in which a genomic feature is translated?
- What is the regulation network involved with a genomic feature of interest?
The website integrates different type of tools for “omics” data analyses:
- A highly interactive genome viewer for display of gene expression arrays, tiling arrays, and RNA-Seq datasets along with proteomics and genomics datasets.
- Expression and protein atlas, which are tools to connect every genomic feature (genes, smallRNAs, antisenseRNAs) or proteins to the most relevant “omics” data.
- A tool to explore gene conservation throughout Listeria strains.
- A co-expression network tool to discover new potential regulation.
Tutorials[edit]
| First hands on Listeriomics | General organization of the website and principal features |
Tools[edit]
| Genomic tools | How-to access all the genomes. |
| Gene tools | How-to access a specific gene and get information about its annotation, its conservation, its transcription or translation. |
| Small RNA tools | How-to access a specific Small RNA |
| Transcriptomic tools | How-to access transcriptomic dataset and display it or perform an analysis with the expression atlas |
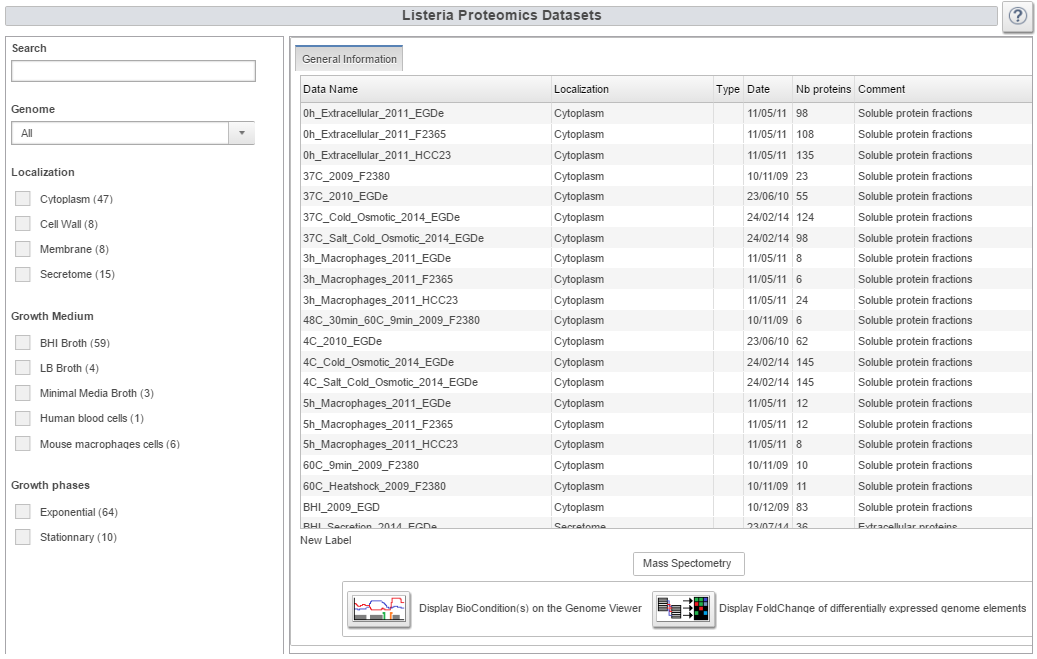
| Proteomic tools | How-to access proteomic dataset and display it or perform an analysis with the protein atlas |
| Co-expression network tools | How-to access to the co-expression network and search for potential co-regulation of genomic feature |
| Submit your datasets | How-to access to the submit section of Listeriomics in which you can inform us of important database update to make |
Viewers[edit]
| Genome viewer | How-to use the genome viewer for visualizing "omics" datasets |
| Transcriptomic HeatMap viewer | How-to use the transcriptomic heatmap viewer and decipher differently expressed genomic features |
| Proteomic HeatMap viewer | How-to use the proteomic heatmap viewer and decipher detected protein in every proteomic experiment |
Database[edit]
The Listeriomics database includes all complete genomes, transcriptomes and proteomes published on Listeria species to date. It allows scientists to analyse dynamically all these datasets in a user-friendly format, and can be used by bio-informaticians as a central database for systems biology analysis.
List of data available:
- 83 Listeria complete genomes
- 492 transcriptome datasets
- 74 proteome datasets
The whole database can be downloaded Here