The aim of the heatmap module is to visualize gene expression fold-changes (shown as log2 values) within different biological conditions.
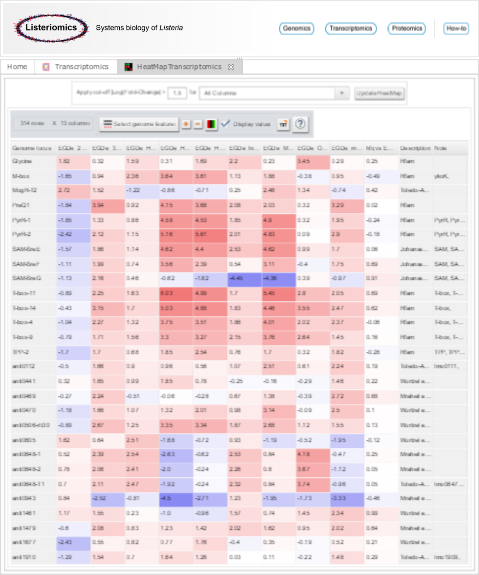 Heatmap visualization of transcriptomics datasets |
Each line of the heatmap corresponds to a gene and each column corresponds to a comparison between two biological conditions
|
Control Panel[edit]
 Control panel for the transcriptomics heatmap visualization |
- Table size: Displays the number of rows and columns.
- Genome features selection: List of genes or non coding RNAs to be displayed in the heatmap viewer can be chosen here. Also, specific lists of elements extracted from key publications can be chosen such as list of internalin genes.
- Zoom in/Zoom out: To increase or decrease the size of the table.
- Change color properties: For fixing the different color displayed in the heatmap. It allows also to select specific colors for some range of values.
- Export in text file: For exporting the table in text format.
|

